wradlib radar data io, visualisation, gridding and gis export
Overview
Within this notebook, we will cover:
Reading radar volume data into xarray based RadarVolume
Examination of RadarVolume and Sweeps
Plotting of sweeps, simple and mapmaking
Gridding and GIS output
Prerequisites
Concepts |
Importance |
Notes |
|---|---|---|
Helpful |
Basic Dataset/DataArray |
|
Helpful |
Basic Plotting |
|
Helpful |
Projections |
|
Helpful |
Raster |
Time to learn: 15 minutes
Imports
import glob
import pathlib
import cartopy
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
from matplotlib import ticker as tick
from osgeo import gdal
import wradlib as wrl
import xradar as xd
Import data into RadarVolume
We have this special case here with Rainbow data where moments are splitted across files. Each file nevertheless consists of all sweeps comprising the volume. We’ll use some special nested ordering to read the files.
fglob = "data/rainbow/meteoswiss/*.vol"
vol = wrl.io.open_rainbow_mfdataset(fglob, combine="by_coords", concat_dim=None)
/home/jgiles/mambaforge/envs/wradlib4/lib/python3.11/site-packages/wradlib/io/rainbow.py:222: FutureWarning: `open_rainbow_mfdataset` is deprecated and will be removed in 2.0. Future development will take place in `xradar`-package.
return open_radar_mfdataset(
Note that we get a warning because this method of loading data will be deprecated in the future in favor of the xradar package. More on alternative loading methods below.
Examine RadarVolume
The RadarVolume is a shallow class which tries to comply to CfRadial2/WMO-FM301, see WMO-CF_Extensions.
The printout of RadarVolume just lists the dimensions and the associated elevations.
display(vol)
<wradlib.RadarVolume>
Dimension(s): (sweep: 10)
Elevation(s): (0.0, 1.3, 2.9, 4.9, 7.3, 10.2, 13.8, 18.2, 23.5, 30.0)
Root Group
The root-group is essentially an overview over the volume, more or less aligned with CfRadial metadata.
vol.root
<xarray.Dataset>
Dimensions: (sweep: 10)
Dimensions without coordinates: sweep
Data variables:
volume_number int64 0
platform_type <U5 'fixed'
instrument_type <U5 'radar'
primary_axis <U6 'axis_z'
time_coverage_start <U20 '2019-10-21T08:24:09Z'
time_coverage_end <U20 '2019-10-21 08:29:34Z'
latitude float64 46.77
longitude float64 6.954
altitude float64 735.0
sweep_group_name (sweep) <U7 'sweep_0' 'sweep_1' ... 'sweep_8' 'sweep_9'
sweep_fixed_angle (sweep) float64 0.0 1.3 2.9 4.9 ... 13.8 18.2 23.5 30.0
Attributes:
version: None
title: None
institution: None
references: None
source: None
history: None
comment: im/exported using wradlib
instrument_name: None
fixed_angle: 0.0Sweep Groups
Sweeps are available in a sequence attached to the RadarVolume object.
swp = vol[0]
display(swp)
<xarray.Dataset>
Dimensions: (azimuth: 360, range: 1400)
Coordinates:
* azimuth (azimuth) float64 0.5 1.5 2.5 3.5 ... 357.5 358.5 359.5
* range (range) float32 25.0 75.0 125.0 ... 6.992e+04 6.998e+04
elevation (azimuth) float64 dask.array<chunksize=(360,), meta=np.ndarray>
rtime (azimuth) datetime64[ns] dask.array<chunksize=(360,), meta=np.ndarray>
sweep_mode <U20 'azimuth_surveillance'
longitude float64 6.954
latitude float64 46.77
altitude float64 735.0
time datetime64[ns] 2019-10-21T08:24:09.041666500
Data variables:
DBZH (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
sweep_number int64 0
prt_mode <U7 'not_set'
follow_mode <U7 'not_set'
sweep_fixed_angle float64 0.0
KDP (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
PHIDP (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
RHOHV (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
VRADH (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
WRADH (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
ZDR (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
Attributes:
fixed_angle: 0.0Inspect Scan Strategy
Considering volume files it’s nice to have an overview over the scan strategy. We can choose some reasonable values for the layout.
# Number of azimuths (rays)
swp["azimuth"].count()
<xarray.DataArray 'azimuth' ()>
array(360)
Coordinates:
sweep_mode <U20 'azimuth_surveillance'
longitude float64 6.954
latitude float64 46.77
altitude float64 735.0
time datetime64[ns] 2019-10-21T08:24:09.041666500# Number of range bins and resolution
swp["range"]
<xarray.DataArray 'range' (range: 1400)>
array([2.5000e+01, 7.5000e+01, 1.2500e+02, ..., 6.9875e+04, 6.9925e+04,
6.9975e+04], dtype=float32)
Coordinates:
* range (range) float32 25.0 75.0 125.0 ... 6.992e+04 6.998e+04
sweep_mode <U20 'azimuth_surveillance'
longitude float64 6.954
latitude float64 46.77
altitude float64 735.0
time datetime64[ns] 2019-10-21T08:24:09.041666500
Attributes:
units: meters
standard_name: projection_range_coordinate
long_name: range_to_measurement_volume
axis: radial_range_coordinate
meters_between_gates: 50.0
spacing_is_constant: true
meters_to_center_of_first_gate: 25.0# Elevations
vol.root.sweep_fixed_angle.values
array([ 0. , 1.3, 2.9, 4.9, 7.3, 10.2, 13.8, 18.2, 23.5, 30. ])
nrays = 360
nbins = 1400
range_res = 50
ranges = np.arange(nbins) * range_res
elevs = vol.root.sweep_fixed_angle.values
sitecoords = (
vol.root.longitude.values.item(),
vol.root.latitude.values.item(),
vol.root.altitude.values.item(),
)
beamwidth = 1.0 # this is unfortunately not in the volume, we need to know this from somewhere else
ax = wrl.vis.plot_scan_strategy(ranges, elevs, sitecoords)
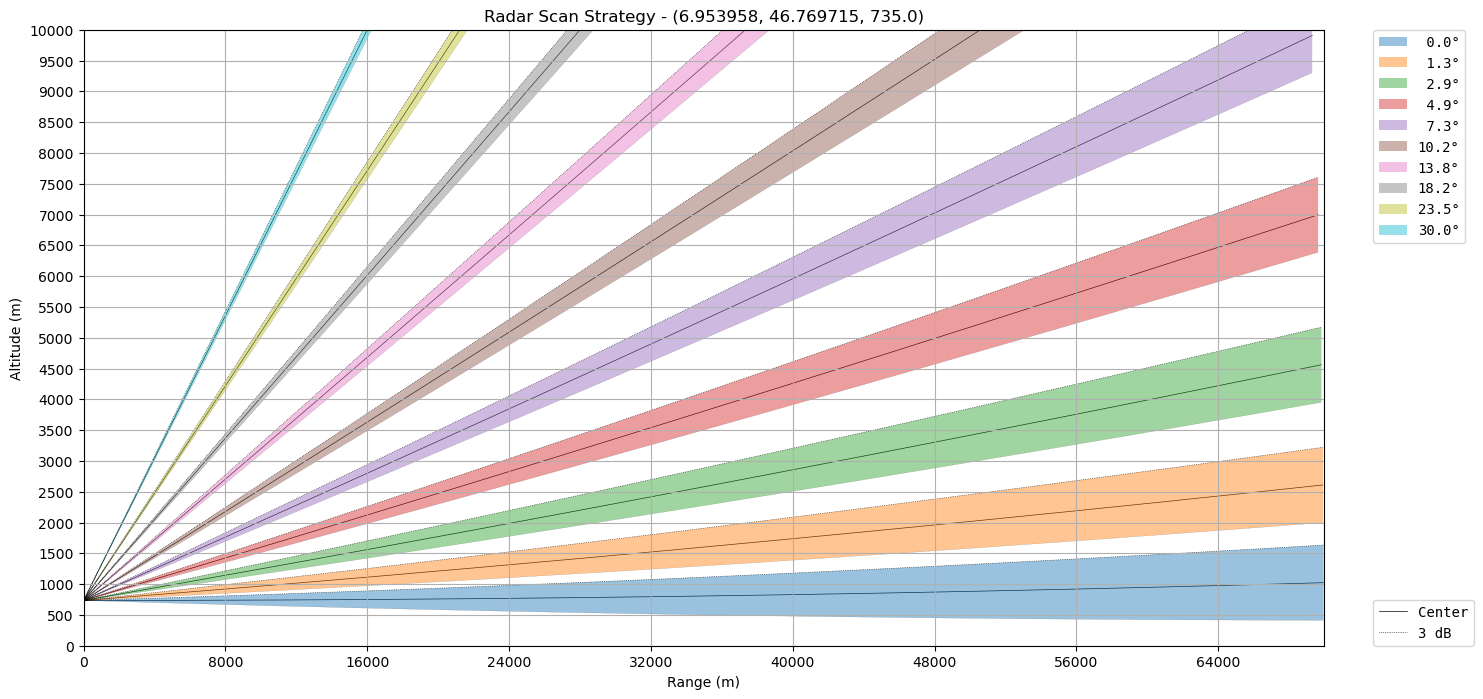
We can plot it on top of the terrain derived from SRTM DEM.
import os
os.environ["WRADLIB_EARTHDATA_BEARER_TOKEN"] = ""
os.environ["WRADLIB_DATA"] = "data/wradlib-data"
ax = wrl.vis.plot_scan_strategy(ranges, elevs, sitecoords, terrain=True)
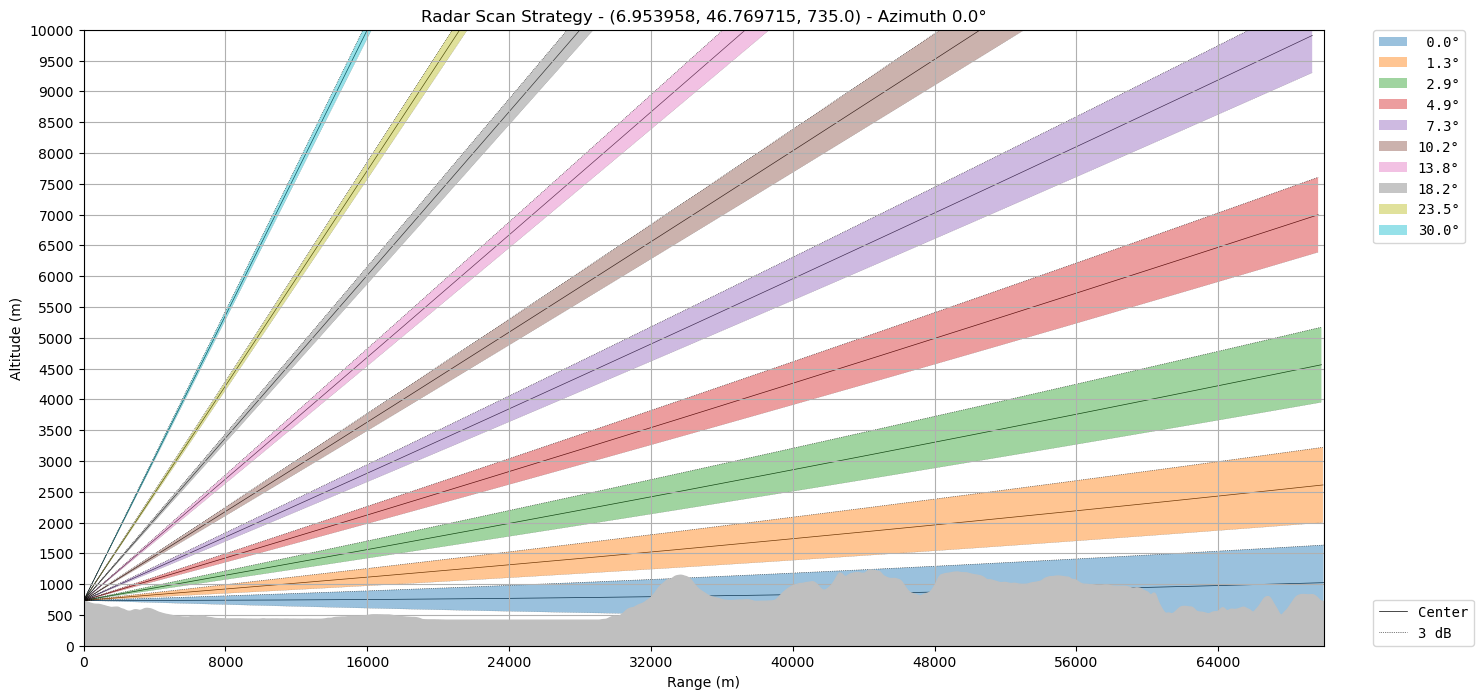
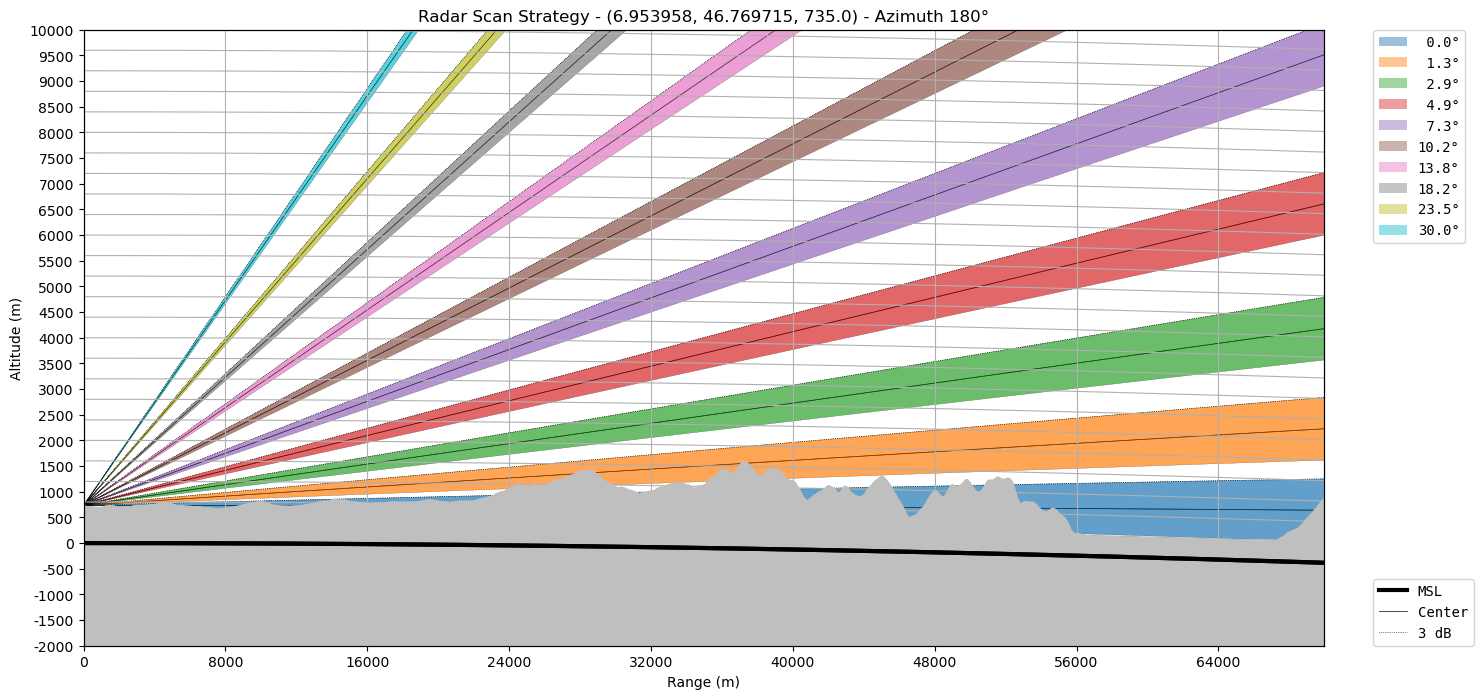
Let’s make the earth go round…
# "cg=True" plots in curvilinear grid and "az=180" plots the 180 degree azimuth
ax = wrl.vis.plot_scan_strategy(
ranges, elevs, sitecoords, cg=True, terrain=True, az=180
)
Plotting Radar Data
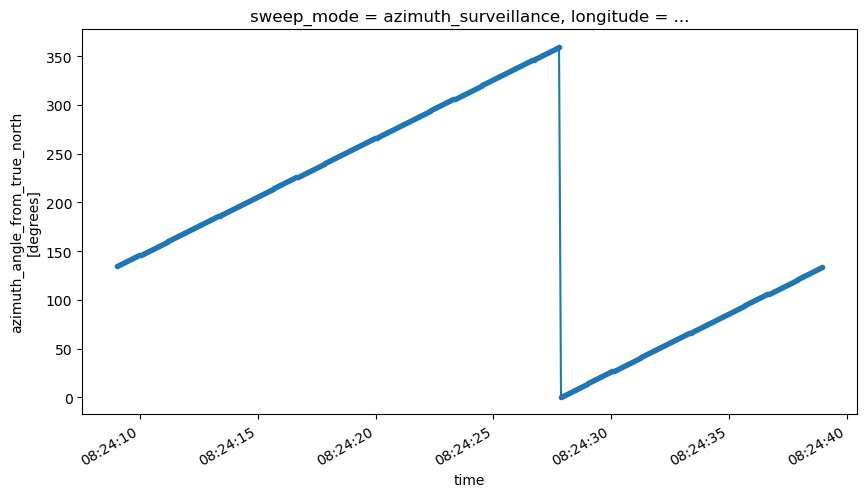
Time vs. Azimuth
fig = plt.figure(figsize=(10, 5))
ax1 = fig.add_subplot(111)
swp.azimuth.sortby("rtime").plot(x="rtime", marker=".")
[<matplotlib.lines.Line2D at 0x7efb97d5c390>]
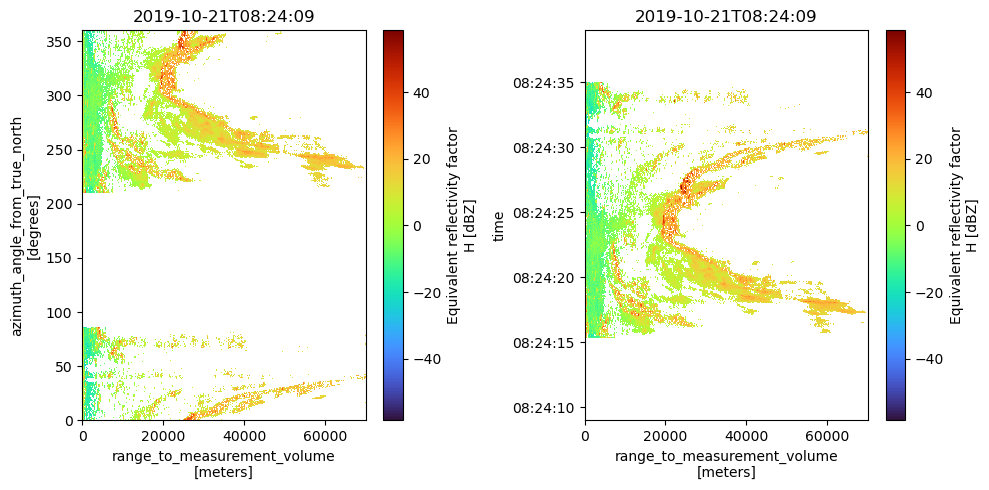
Range vs. Azimuth/Time
fig = plt.figure(figsize=(10, 5))
ax1 = fig.add_subplot(121)
swp.DBZH.plot(cmap="turbo", ax=ax1)
ax1.set_title(f"{swp.time.values.astype('M8[s]')}")
ax2 = fig.add_subplot(122)
swp.DBZH.sortby("rtime").plot(y="rtime", cmap="turbo", ax=ax2)
ax2.set_title(f"{swp.time.values.astype('M8[s]')}")
plt.tight_layout()
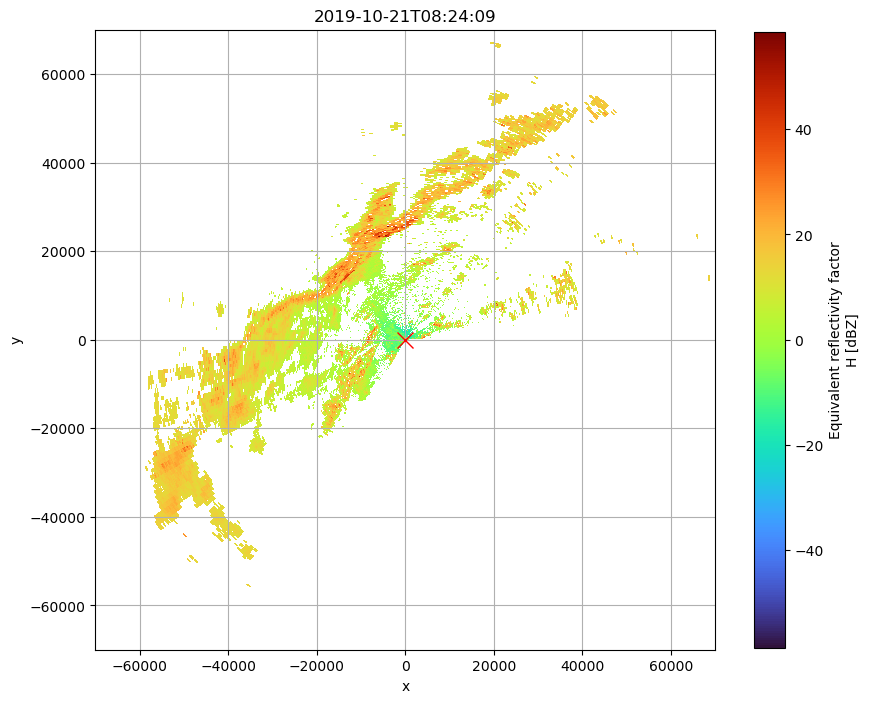
Georeferenced as Plan Position Indicator
fig = plt.figure(figsize=(10, 10))
ax1 = fig.add_subplot(111)
swp.DBZH.pipe(wrl.georef.georeference_dataset).plot(
x="x", y="y", ax=ax1, cmap="turbo", cbar_kwargs=dict(shrink=0.8)
)
ax1.plot(0, 0, "rx", markersize=12)
ax1.set_title(f"{swp.time.values.astype('M8[s]')}")
ax1.grid()
ax1.set_aspect("equal")
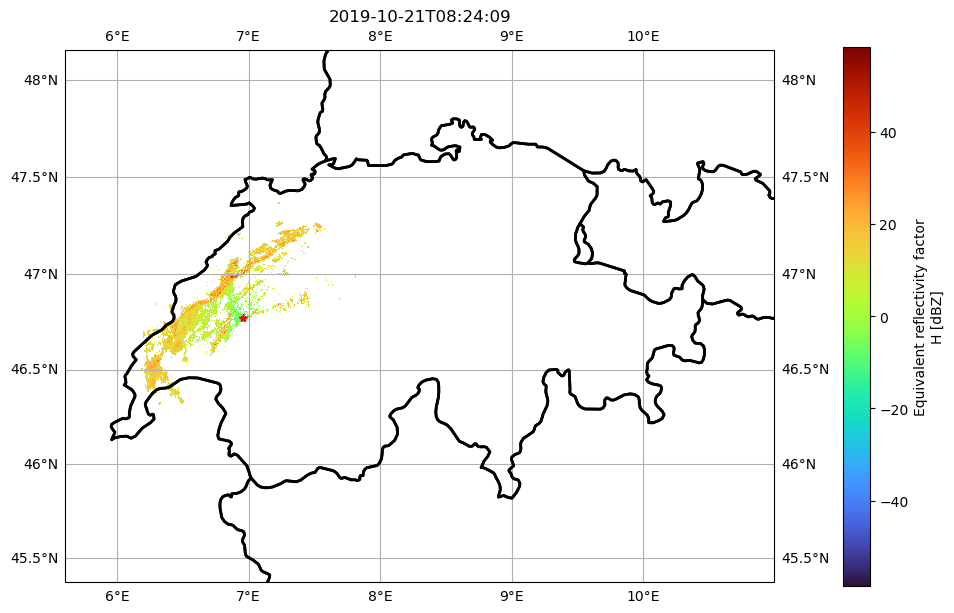
Basic MapMaking with cartopy
The data will be georeferenced as Azimuthal Equidistant Projection centered at the radar. For the map projection we will use Mercator.
map_trans = ccrs.AzimuthalEquidistant(
central_latitude=swp.latitude.values, central_longitude=swp.longitude.values
)
map_proj = ccrs.Mercator(central_longitude=swp.longitude.values)
def plot_borders(ax):
borders = cfeature.NaturalEarthFeature(
category="cultural", name="admin_0_countries", scale="10m", facecolor="none"
)
ax.add_feature(borders, edgecolor="black", lw=2, zorder=4)
fig = plt.figure(figsize=(10, 8))
ax = fig.add_subplot(111, projection=map_proj)
cbar_kwargs = dict(shrink=0.7, pad=0.075)
pm = swp.DBZH.pipe(wrl.georef.georeference_dataset).plot(
ax=ax, x="x", y="y", cbar_kwargs=cbar_kwargs, cmap="turbo", transform=map_trans
)
plot_borders(ax)
ax.gridlines(draw_labels=True)
ax.plot(
swp.longitude.values, swp.latitude.values, transform=map_trans, marker="*", c="r"
)
ax.set_title(f"{swp.time.values.astype('M8[s]')}")
ax.set_xlim(-15e4, 45e4)
ax.set_ylim(565e4, 610e4)
plt.tight_layout()
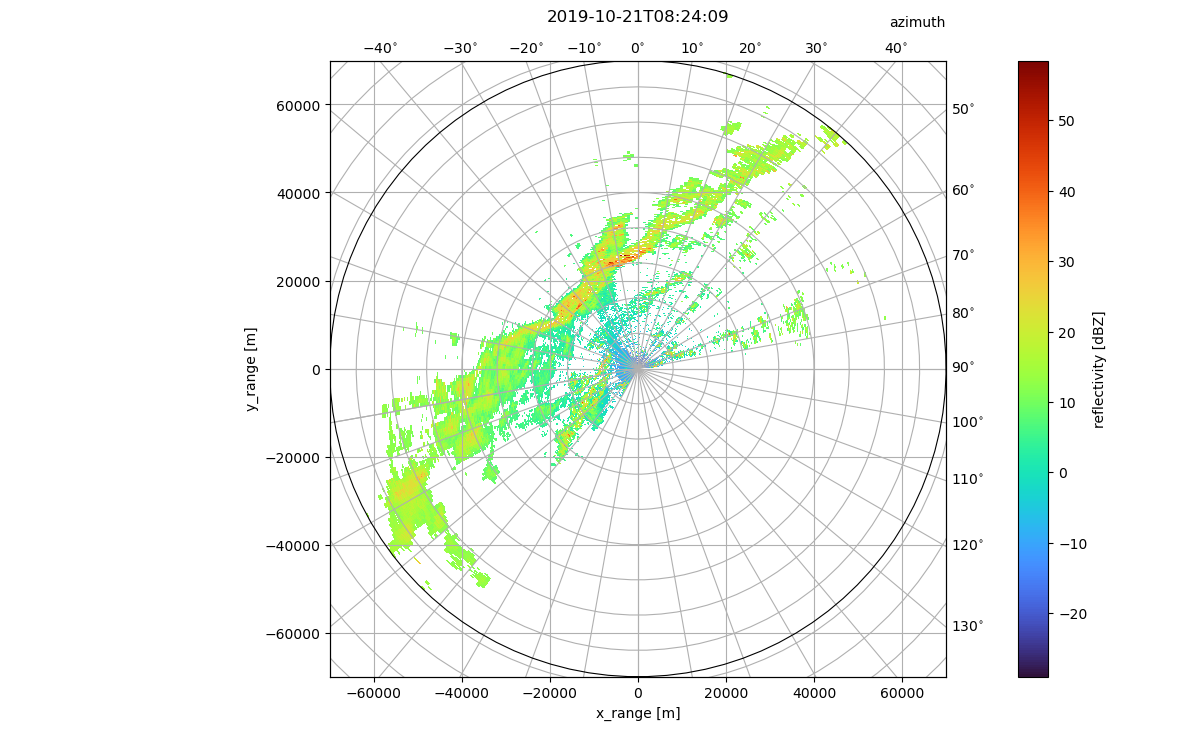
Plot on curvelinear grid
For Xarray DataArrays wradlib uses a so-called accessor (wradlib). To plot on curvelinear grids projection has to be set to cg, which uses the matplotlib AXISARTIS namespace.
fig = plt.figure(figsize=(14, 8))
pm = swp.DBZH.pipe(wrl.georef.georeference_dataset).wradlib.plot(
proj="cg", fig=fig, cmap="turbo"
)
ax = plt.gca()
# apply eye-candy
caax = ax.parasites[0]
paax = ax.parasites[1]
ax.parasites[1].set_aspect("equal")
t = plt.title(f"{vol[0].time.values.astype('M8[s]')}", y=1.05)
cbar = plt.colorbar(pm, pad=0.075, ax=paax)
caax.set_xlabel("x_range [m]")
caax.set_ylabel("y_range [m]")
plt.text(1.0, 1.05, "azimuth", transform=caax.transAxes, va="bottom", ha="right")
cbar.set_label("reflectivity [dBZ]")
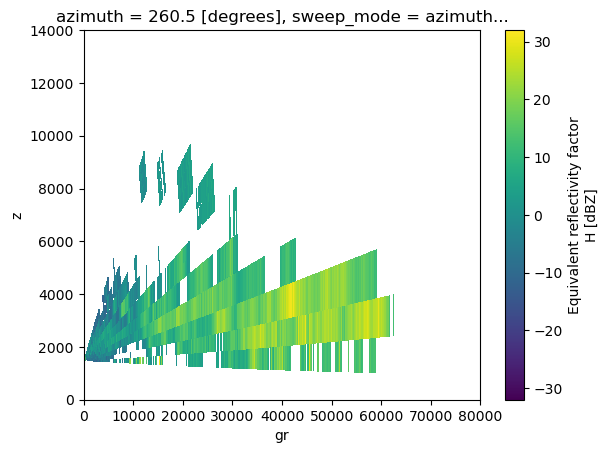
Making a vertical cut of the volume (fake RHI)
New function cross_section_ppi since Wradlib 1.18. Similar to Py-ART’s cross_section_ppi
The volume elements need to have time dimension even if it is a single timestep (because how utility functions in wradlib work), then we add time dimension first:
# we create an empty volume
vol2 = wrl.io.xarray.RadarVolume()
# we take each element in vol, add time dimension and append it to vol2
for vv in vol:
vol2.append(vv.expand_dims("time"))
We apply the function to extract a cross section of the volume at a certain azimuth:
# Extract a single azimuth
azimuth = 260.5
rec_rhi = wrl.util.cross_section_ppi(vol2, azimuth)
Plot the result:
rec_rhi.DBZH[0].plot(cmap='viridis', x="gr", y="z", ylim=(0,14000), xlim=(0, 80000))
<matplotlib.collections.QuadMesh at 0x7efb8c984f10>
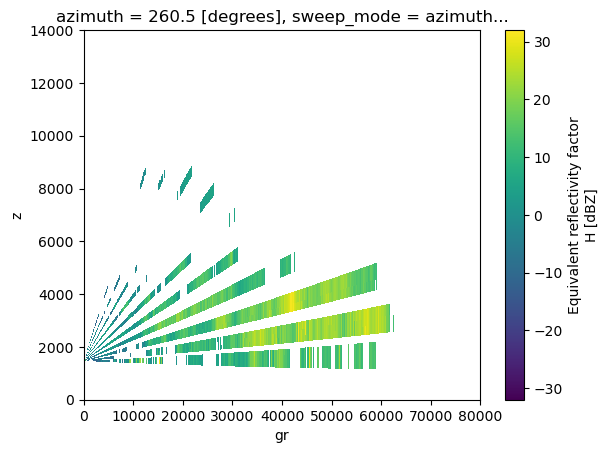
We can add the option real_beams=True, which will generate “fake” empty beams to produce a dataset that, when plotted, represents the real beam width (be default beam width = 1 degree)
bw = 1 # beam width in degrees, this will depend on the radar
rec_rhi = wrl.util.cross_section_ppi(vol2, azimuth, real_beams=True, bw=bw)
# Plot result
rec_rhi.DBZH[0].plot(cmap='viridis', x="gr", y="z", ylim=(0,14000), xlim=(0, 80000))
<matplotlib.collections.QuadMesh at 0x7efb8f8e7ed0>
We can extract two opposing azimuths and plot both left and right of the radar:
azimuths = [azimuth, azimuth-180]
rec_rhi_2 = wrl.util.cross_section_ppi(vol2, azimuths)
rec_rhi0 = rec_rhi_2.isel(azimuth=0)
rec_rhi180 = rec_rhi_2.isel(azimuth=1)
# we have to invert the gr and range coordinates in one of them so that they are opposite to each other
rec_rhi180.coords["gr"] = rec_rhi180.coords["gr"]*-1
rec_rhi180.coords["range"] = rec_rhi180.coords["range"]*-1
# PLot
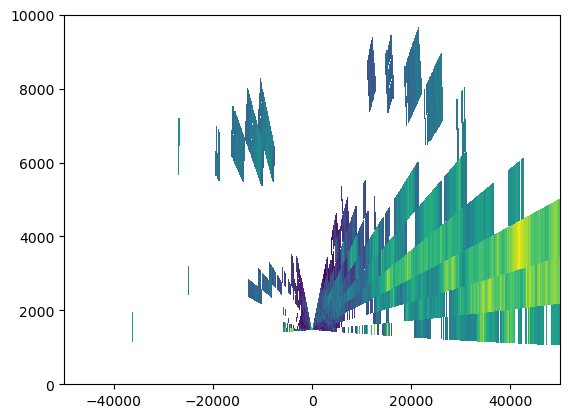
plt.pcolormesh(rec_rhi0.gr, rec_rhi0.z, rec_rhi0.DBZH[0], cmap='viridis')
plt.pcolormesh(rec_rhi180.gr, rec_rhi180.z, rec_rhi180.DBZH[0], cmap='viridis')
plt.xlim(-50000, 50000)
plt.ylim(0, 10000)
/tmp/ipykernel_15797/1949423905.py:2: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(rec_rhi0.gr, rec_rhi0.z, rec_rhi0.DBZH[0], cmap='viridis')
/tmp/ipykernel_15797/1949423905.py:3: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
plt.pcolormesh(rec_rhi180.gr, rec_rhi180.z, rec_rhi180.DBZH[0], cmap='viridis')
(0.0, 10000.0)
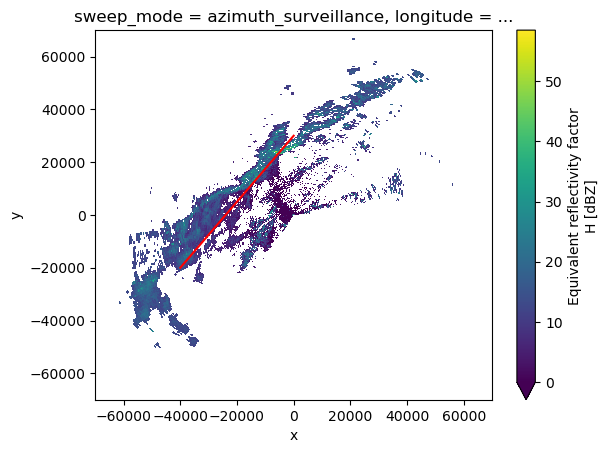
We can make an arbitrary cut by providing two points through where a line of values will be extracted
# Choose two points (in meters in the coordinates x,y of the georeferenced data)
vol0 = wrl.georef.georeference_dataset(vol2[0])
p1 = (-40000, -20000)
p2 = (0, 30000)
# Plot the chosen line over a PPI
vol0.DBZH[0].plot(x="x", y="y", vmin=0, cmap="viridis")
plt.plot([p1[0], p2[0]], [p1[1], p2[1]], c="red")
[<matplotlib.lines.Line2D at 0x7efb8f6c1090>]
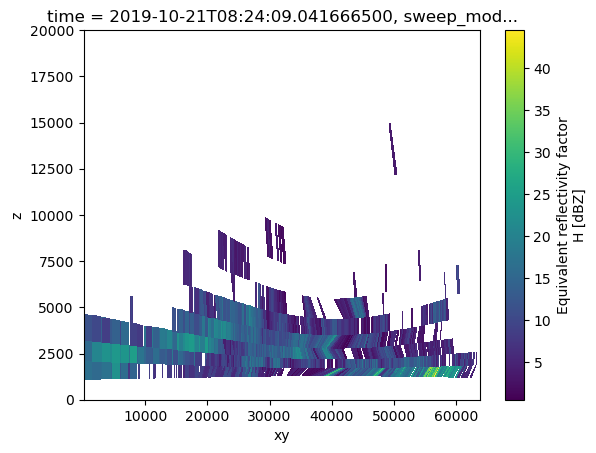
# Extract cross section and plot result
rec_rhi = wrl.util.cross_section_ppi(vol2, (p1,p2))
# We have to use the new coordinate "xy" (meters along the line from p1 to p2) for plotting
rec_rhi.DBZH[0].plot(cmap="viridis", x="xy", y="z", ylim=(0,20000))
<matplotlib.collections.QuadMesh at 0x7efb8cb66850>
ODIM_H5 format export and import
Export to ODIM_H5
To save the file, we need to pass a “source” parameter compliant with the format standard:
http://eumetnet.eu/wp-content/uploads/2017/01/OPERA_hdf_description_2014.pdf (page 11)
for example, source=“RAD:xxxx”
# I don't know the exact code for this radar so let's just put zeros
vol.to_odim("test_odim_vol.h5", source="RAD:0000")
Import from ODIM_H5
vol2 = wrl.io.open_odim_dataset("test_odim_vol.h5")
display(vol2)
/home/jgiles/mambaforge/envs/wradlib4/lib/python3.11/site-packages/wradlib/io/hdf.py:92: FutureWarning: `open_odim_dataset` functionality has been moved to `xradar`-package and will be removed in 2.0. Use `open_odim_datatree` from `xradar`-package.
return open_radar_dataset(filename_or_obj, engine=OdimBackendEntrypoint, **kwargs)
<wradlib.RadarVolume>
Dimension(s): (sweep: 10)
Elevation(s): (0.0, 1.3, 2.9, 4.9, 7.3, 10.2, 13.8, 18.2, 23.5, 30.0)
display(vol2[0])
<xarray.Dataset>
Dimensions: (azimuth: 360, range: 1400)
Coordinates:
* azimuth (azimuth) float32 0.5 1.5 2.5 3.5 ... 357.5 358.5 359.5
elevation (azimuth) float32 ...
rtime (azimuth) datetime64[ns] ...
* range (range) float32 25.0 75.0 125.0 ... 6.992e+04 6.998e+04
sweep_mode <U20 ...
longitude float64 ...
latitude float64 ...
altitude float64 ...
time datetime64[ns] 2019-10-21T08:24:09.041666816
Data variables:
DBZH (azimuth, range) float32 ...
KDP (azimuth, range) float32 ...
PHIDP (azimuth, range) float32 ...
RHOHV (azimuth, range) float32 ...
VRADH (azimuth, range) float32 ...
WRADH (azimuth, range) float32 ...
ZDR (azimuth, range) float32 ...
sweep_number int64 ...
prt_mode <U7 ...
follow_mode <U7 ...
sweep_fixed_angle float64 0.0
Attributes:
fixed_angle: 0.0Import with xarray backends
This is now the recommended way of opening files!
We can facilitate the xarray backend’s which xradar (previosly wradlib) provides for the different readers. The xarray backends are capable of loading data into a single Dataset.
Open single files
The simplest case can only open one file
ds = xr.open_dataset("test_odim_vol.h5", engine="odim")
display(ds)
<xarray.Dataset>
Dimensions: (azimuth: 360, range: 1400)
Coordinates:
* azimuth (azimuth) float32 0.5 1.5 2.5 3.5 ... 357.5 358.5 359.5
elevation (azimuth) float32 ...
time (azimuth) datetime64[ns] ...
* range (range) float32 25.0 75.0 125.0 ... 6.992e+04 6.998e+04
longitude float64 ...
latitude float64 ...
altitude float64 ...
Data variables:
DBZH (azimuth, range) float32 ...
KDP (azimuth, range) float32 ...
PHIDP (azimuth, range) float32 ...
RHOHV (azimuth, range) float32 ...
VRADH (azimuth, range) float32 ...
WRADH (azimuth, range) float32 ...
ZDR (azimuth, range) float32 ...
sweep_mode <U20 ...
sweep_number int64 ...
prt_mode <U7 ...
follow_mode <U7 ...
sweep_fixed_angle float64 ...Open multiple files
Here we need to specify the group, which in case of rainbow files is given by the group number.
ds = xr.open_mfdataset(fglob, engine="rainbow", group="sweep_0", combine="by_coords")
display(ds)
<xarray.Dataset>
Dimensions: (azimuth: 360, range: 1400)
Coordinates:
* azimuth (azimuth) float64 0.4999 1.505 2.505 ... 358.5 359.5
elevation (azimuth) float64 dask.array<chunksize=(360,), meta=np.ndarray>
* range (range) float32 25.0 75.0 125.0 ... 6.992e+04 6.998e+04
time (azimuth) datetime64[ns] dask.array<chunksize=(360,), meta=np.ndarray>
longitude float64 6.954
latitude float64 46.77
altitude float64 735.0
Data variables:
DBZH (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
sweep_mode <U20 'azimuth_surveillance'
sweep_number int64 0
prt_mode <U7 'not_set'
follow_mode <U7 'not_set'
sweep_fixed_angle float64 0.0
KDP (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
PHIDP (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
RHOHV (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
VRADH (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
WRADH (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>
ZDR (azimuth, range) float32 dask.array<chunksize=(360, 1400), meta=np.ndarray>If we want to have everything in a radar volume, we have to open all sweeps and build it manually
# we create an empty volume
vol = wrl.io.xarray.RadarVolume()
# we take each element in vol, add time dimension and append it to vol2
for n in np.arange(10):
vol.append(xr.open_mfdataset(fglob, engine="rainbow", group="sweep_"+str(n), combine="by_coords"))
vol
<wradlib.RadarVolume>
Dimension(s): (sweep: 10)
Elevation(s): (0.0, 1.3, 2.9, 4.9, 7.3, 10.2, 13.8, 18.2, 23.5, 30.0)
Gridding and Export to GIS formats
get coordinates from source Dataset with given projection
calculate target coordinates
grid using wradlib interpolator
export to single band geotiff
use GDAL CLI tools to convert to grayscaled/paletted PNG
def get_target_grid(ds, nb_pixels):
xgrid = np.linspace(ds.x.min(), ds.x.max(), nb_pixels, dtype=np.float32)
ygrid = np.linspace(ds.y.min(), ds.y.max(), nb_pixels, dtype=np.float32)
grid_xy_raw = np.meshgrid(xgrid, ygrid)
grid_xy_grid = np.dstack((grid_xy_raw[0], grid_xy_raw[1]))
return xgrid, ygrid, grid_xy_grid
def get_target_coordinates(grid):
grid_xy = np.stack((grid[..., 0].ravel(), grid[..., 1].ravel()), axis=-1)
return grid_xy
def get_source_coordinates(ds):
xy = np.stack((ds.x.values.ravel(), ds.y.values.ravel()), axis=-1)
return xy
def coordinates(da, proj, res=100):
# georeference single sweep
da = da.pipe(wrl.georef.georeference_dataset, proj=proj)
# get source coordinates
src = get_source_coordinates(da)
# create target grid
xgrid, ygrid, trg = get_target_grid(da, res)
return src, trg
def moment_to_gdal(da, trg_grid, driver, ext, path="", proj=None):
# use wgs84 pseudo mercator if no projection is given
if proj is None:
proj = wrl.georef.epsg_to_osr(3857)
t = da.time[0].values.astype("M8[s]").astype("O")
outfilename = f"gridded_{da.name}_{t:%Y%m%d}_{t:%H%M%S}"
outfilename = os.path.join(path, outfilename)
f = pathlib.Path(outfilename)
f.unlink(missing_ok=True)
res = ip_near(da.values.ravel(), maxdist=1000).reshape(
(len(trg_grid[0]), len(trg_grid[1]))
)
data, xy = wrl.georef.set_raster_origin(res, trg_grid, "upper")
ds = wrl.georef.create_raster_dataset(data, xy, projection=proj)
wrl.io.write_raster_dataset(outfilename + ext, ds, driver)
Coordinates
%%time
epsg_code = 2056
proj = wrl.georef.epsg_to_osr(epsg_code)
src, trg = coordinates(ds, proj, res=1400)
CPU times: user 590 ms, sys: 46.6 ms, total: 637 ms
Wall time: 641 ms
Interpolator
%%time
ip_near = wrl.ipol.Nearest(src, trg.reshape(-1, trg.shape[-1]), remove_missing=7)
CPU times: user 2.09 s, sys: 381 ms, total: 2.47 s
Wall time: 2.48 s
Gridding and Export
%%time
moment_to_gdal(ds.DBZH, trg, "GTiff", ".tif", proj=proj)
CPU times: user 326 ms, sys: 85.2 ms, total: 411 ms
Wall time: 421 ms
GDAL info on created GeoTiff
!gdalinfo gridded_DBZH_20191021_082427.tif
Driver: GTiff/GeoTIFF
Files: gridded_DBZH_20191021_082427.tif
Size is 1400, 1400
Coordinate System is:
PROJCRS["CH1903+ / LV95",
BASEGEOGCRS["CH1903+",
DATUM["CH1903+",
ELLIPSOID["Bessel 1841",6377397.155,299.1528128,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4150]],
CONVERSION["Swiss Oblique Mercator 1995",
METHOD["Hotine Oblique Mercator (variant B)",
ID["EPSG",9815]],
PARAMETER["Latitude of projection centre",46.9524055555556,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8811]],
PARAMETER["Longitude of projection centre",7.43958333333333,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8812]],
PARAMETER["Azimuth of initial line",90,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8813]],
PARAMETER["Angle from Rectified to Skew Grid",90,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8814]],
PARAMETER["Scale factor on initial line",1,
SCALEUNIT["unity",1],
ID["EPSG",8815]],
PARAMETER["Easting at projection centre",2600000,
LENGTHUNIT["metre",1],
ID["EPSG",8816]],
PARAMETER["Northing at projection centre",1200000,
LENGTHUNIT["metre",1],
ID["EPSG",8817]]],
CS[Cartesian,2],
AXIS["(E)",east,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["(N)",north,
ORDER[2],
LENGTHUNIT["metre",1]],
USAGE[
SCOPE["Cadastre, engineering survey, topographic mapping (large and medium scale)."],
AREA["Liechtenstein; Switzerland."],
BBOX[45.82,5.96,47.81,10.49]],
ID["EPSG",2056]]
Data axis to CRS axis mapping: 1,2
Origin = (2492961.000000000000000,1249970.687500000000000)
Pixel Size = (100.000000000000000,-100.125000000000000)
Metadata:
AREA_OR_POINT=Area
Image Structure Metadata:
INTERLEAVE=BAND
Corner Coordinates:
Upper Left ( 2492961.000, 1249970.688) ( 6d 1'17.65"E, 47d23'35.61"N)
Lower Left ( 2492961.000, 1109795.688) ( 6d 3'15.75"E, 46d 7'56.52"N)
Upper Right ( 2632961.000, 1249970.688) ( 7d52'34.61"E, 47d24' 3.98"N)
Lower Right ( 2632961.000, 1109795.688) ( 7d51'58.24"E, 46d 8'24.24"N)
Center ( 2562961.000, 1179883.188) ( 6d57'16.56"E, 46d46'13.43"N)
Band 1 Block=1400x1 Type=Float32, ColorInterp=Gray
NoData Value=-9999
Translate exported GeoTiff to grayscale PNG
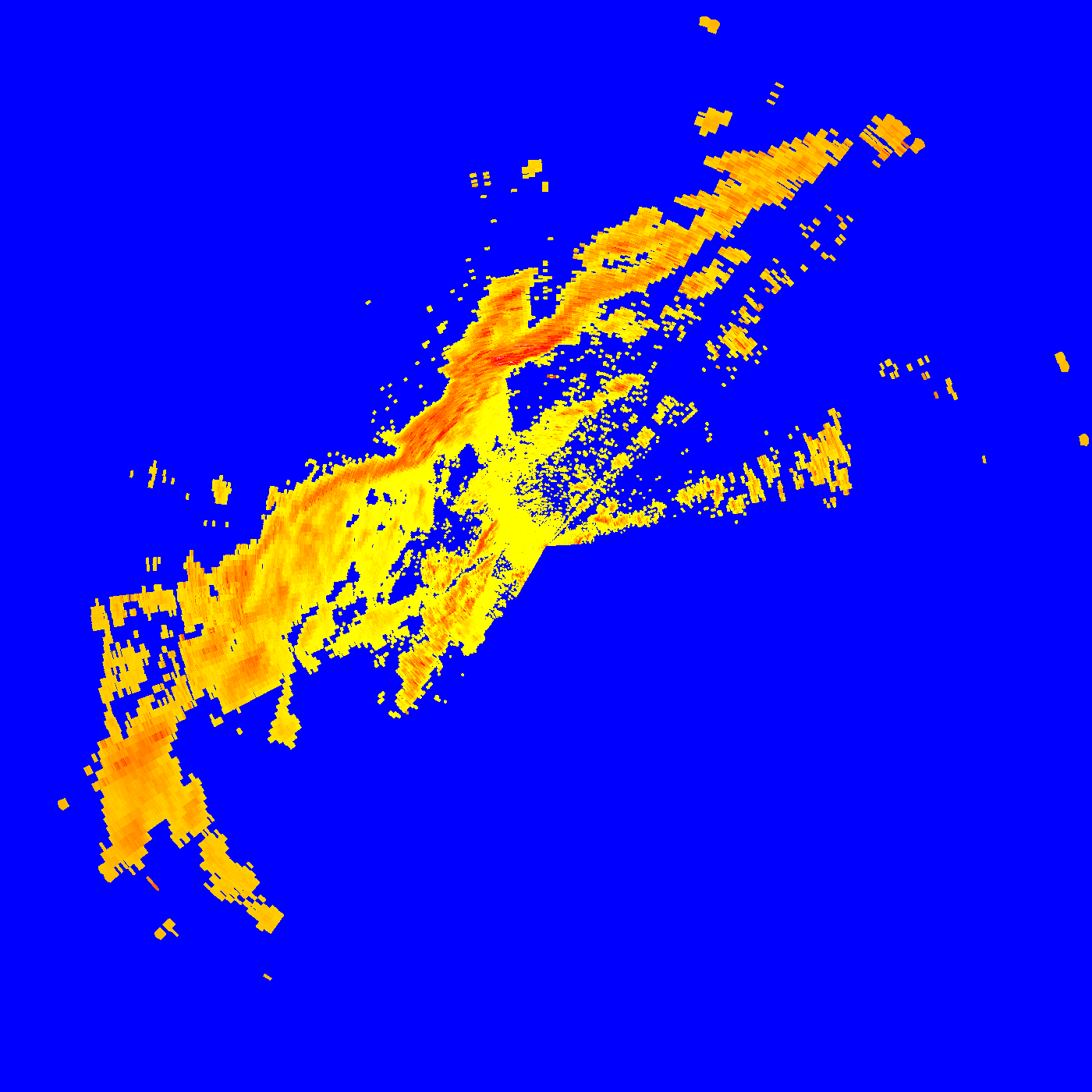
!gdal_translate -of PNG -ot Byte -scale -30. 60. 0 255 gridded_DBZH_20191021_082427.tif grayscale.png
Input file size is 1400, 1400
Warning 1: for band 1, nodata value has been clamped to 0, the original value being out of range.
0...10...20...30...40...50...60...70...80...90...100 - done.
Apply colortable to PNG
with open("colors.txt", "w") as f:
f.write("0 blue\n")
f.write("50 yellow\n")
f.write("100 yellow\n")
f.write("150 orange\n")
f.write("200 red\n")
f.write("250 white\n")
Display exported PNG’s
!gdaldem color-relief grayscale.png colors.txt paletted.png
0...10...20...30...40...50...60...70...80...90...100 - done.
Import with Xarray, rasterio backend
with xr.open_dataset("gridded_DBZH_20191021_082427.tif", engine="rasterio") as ds_grd:
display(ds_grd)
ds_grd.band_data.plot(cmap="turbo")
/home/jgiles/mambaforge/envs/wradlib4/lib/python3.11/site-packages/pyproj/crs/_cf1x8.py:514: UserWarning: angle from rectified to skew grid parameter lost in conversion to CF
warnings.warn(
<xarray.Dataset>
Dimensions: (band: 1, x: 1400, y: 1400)
Coordinates:
* band (band) int64 1
* x (x) float64 2.493e+06 2.493e+06 ... 2.633e+06 2.633e+06
* y (y) float64 1.25e+06 1.25e+06 1.25e+06 ... 1.11e+06 1.11e+06
spatial_ref int64 ...
Data variables:
band_data (band, y, x) float32 ...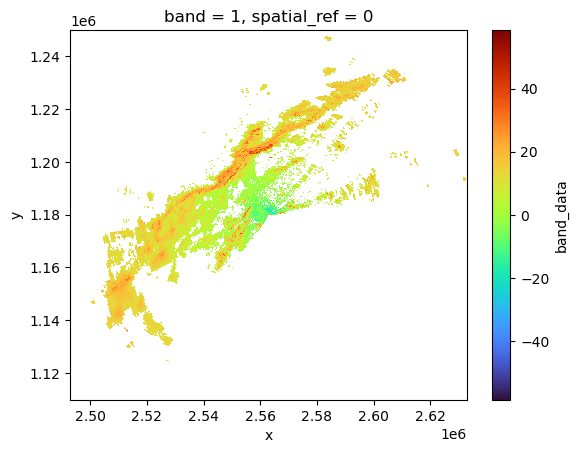
xradar
Import with xradar
We can use xradar to open the rainbow files, but only one at the time. Let’s open the first one:
files = glob.glob(fglob)
vol_xd = xd.io.open_rainbow_datatree(files[0])
vol_xd
<xarray.DatasetView>
Dimensions: ()
Data variables:
volume_number int64 0
platform_type <U5 'fixed'
instrument_type <U5 'radar'
time_coverage_start <U20 '2019-10-21T08:24:09Z'
time_coverage_end <U20 '2019-10-21T08:29:33Z'
longitude float64 6.954
altitude float64 735.0
latitude float64 46.77
Attributes:
Conventions: None
version: None
title: None
institution: None
references: None
source: None
history: None
comment: im/exported using xradar
instrument_name: Nonevol_xd["sweep_9"]
<xarray.DatasetView>
Dimensions: (azimuth: 360, range: 1400)
Coordinates:
* azimuth (azimuth) float64 0.4999 1.502 2.497 ... 358.5 359.5
elevation (azimuth) float64 ...
* range (range) float32 25.0 75.0 125.0 ... 6.992e+04 6.998e+04
time (azimuth) datetime64[ns] 2019-10-21T08:29:27.541666499...
longitude float64 ...
latitude float64 ...
altitude float64 ...
Data variables:
KDP (azimuth, range) float32 ...
sweep_mode <U20 ...
sweep_number int64 ...
prt_mode <U7 ...
follow_mode <U7 ...
sweep_fixed_angle float64 ...We can transform it to an xarray Dataset like this:
vol_xd["sweep_9"].to_dataset()
<xarray.Dataset>
Dimensions: (azimuth: 360, range: 1400)
Coordinates:
* azimuth (azimuth) float64 0.4999 1.502 2.497 ... 358.5 359.5
elevation (azimuth) float64 ...
* range (range) float32 25.0 75.0 125.0 ... 6.992e+04 6.998e+04
time (azimuth) datetime64[ns] 2019-10-21T08:29:27.541666499...
longitude float64 ...
latitude float64 ...
altitude float64 ...
Data variables:
KDP (azimuth, range) float32 ...
sweep_mode <U20 ...
sweep_number int64 ...
prt_mode <U7 ...
follow_mode <U7 ...
sweep_fixed_angle float64 ...You can then open several files into datatrees, transform them to xarray Dataset and merge them into a single Dataset.
Open other formats
armor = "../../data/qc/uah-armor/cfrad.20080411_182230.747_to_20080411_182629.530_ARMOR_SUR.nc"
armor_swp = xr.open_dataset(armor, group="sweep_0", engine="cfradial1")
armor_swp
<xarray.Dataset>
Dimensions: (time: 4319, range: 992, azimuth: 360)
Coordinates:
* time (time) datetime64[ns] 2008-04-11T18:22:30.7469...
* range (range) float32 1e+03 1.125e+03 ... 1.249e+05
* azimuth (azimuth) float32 0.02747 1.052 ... 358.0 359.1
elevation (azimuth) float32 ...
latitude (time) float64 ...
longitude (time) float64 ...
altitude (time) float64 ...
Data variables: (12/47)
sweep_number float64 ...
sweep_mode |S32 ...
prt_mode |S32 ...
follow_mode |S32 ...
sweep_fixed_angle float32 ...
ray_start_range (azimuth) float32 ...
... ...
PHIDP (azimuth, range) float32 ...
RHOHV (azimuth, range) float32 ...
WIDTH (azimuth, range) float32 ...
VEL (azimuth, range) float32 ...
REF (azimuth, range) float32 ...
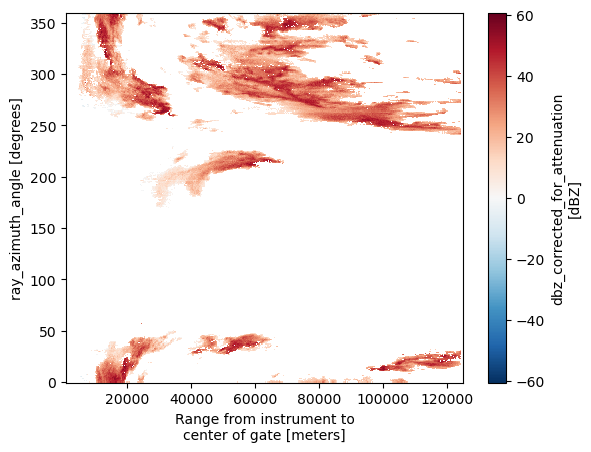
VR (azimuth, range) float32 ...armor_swp["DBZ"].plot()
<matplotlib.collections.QuadMesh at 0x7efb8c3f7ed0>